Development of methods for the genomic analysis of microorganisms in bioaerosols
DNA-Sequencing, a key method in molecular biology and bioengineering, is a fundamental pillar to modern biotechnology and molecular diagnostics. Of age and well established are first (i.e. Sanger) and second (i.e. next-gen) generation sequencing methods. Both share comparatively high up-front and running costs, as well as comparatively short read-lengths. The latter are negatively affecting downstream data-analyses. The advent of third generation or single molecule sequencing technology (i.e. nanopore) offers great potential to decrease costs and increasing accessibility, while improving read lengths massively. The latter benefits downstream data-analyses.
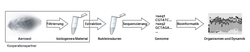
Bioaerosols are ubiquitous and very complex airborne particles of biological origin. They contain parts of and indeed complete microorganisms, e.g. bacteria and fungi, or viruses. These may be harmful to the health of human beings. Subsequently they are relevant in the context of environmental and occupational health, but also within the scope of biosafety, -security and environmental protection. The accompanying diagnostics are elaborate and to date limited with respect to quantitative and qualitative approaches.
Aim of research is the development and introduction of a methodology enabling molecular surveillance of the microbial load of extracted air from e.g. animal husbandry (farms) or bioreactors (biogas plants). The reproducible and quantitative assessment generated offers a knowledge basis with respect to occupational and environmental safety.
The pipeline developed within the scope of this project is based on nanopore sequencing and circumvents drawbacks and limitations of currently established first and second generation sequencers (high up-front and running costs, complex downstream data analyses), and provides substantial enhancement for bioaerosol surveillance.
Supervisor: Professor Dr. Röbbe Wünschiers, Chair of Biochemistry and Molecular Biology, Mittweida UAS
Supervisor: Professor Dr. Michael Göttfert, Chair of Molecular Genetics, TU Dresden
In collaboration with:
Federal Institute for Occupational Safety and Health
Saxon Ministry for Environment and Agriculture
Publications
Leidenfrost RM, Pöther D, Jäckel U, Wünschiers R, (2020) Benchmarking the MinION: Evaluating long reads for microbial profiling. Sci Rep. doi: 10.1038/s41598-020-61989-x
Conference and conference contributions (selection)
Reading Plant ITS2-Sequences with Nanopores and Polymerases: First Insights to Pros and Cons (Poster)
Leidenfrost, Robert; Bänsch, Svenja; Prudnikow, Lisa; Westphal, Catrin; Wünschiers,
Röbbe
15th Gatersleben Research Conference on Applied Bioinformatics in Crops
Gatersleben/Germany, March 18-20, 2019
An introduction to nanopore sequencing (Talk)
Leidenfrost, Robert
ICOLE - Workshop for Computational Biology, Scheduling, and Machine Learning
Lessach/Austria, September 24-28, 2018
Viability of long noisy reads for (meta)genomics applications (Talk)
Leidenfrost, Robert
Mittelerde-Meeting 2018 – 3rd Central German Meeting on Bioinformatics
Mittweida/Germany, June 14-15, 2018
Entwicklung von Methoden für die genombasierte Analyse von Mikroorganismen in
Bioaerosolen (Poster)
Robert Leidenfrost & Röbbe Wünschiers
VDI Expertenforum Bioaerosole – Von der Messung zur Bewertung
Berlin/Germany; September 26-27, 2017